A collection of informal reviews of papers I find interesting — mostly in the protein
structure prediction, protein design, and protein language model space. These are from
Sergey Ovchinnikov's lab
and related groups. Just my thoughts, nothing too formal.
Protein Diffusion Models as Statistical Potentials
Roney, Ou, Ovchinnikov · bioRxiv 2025
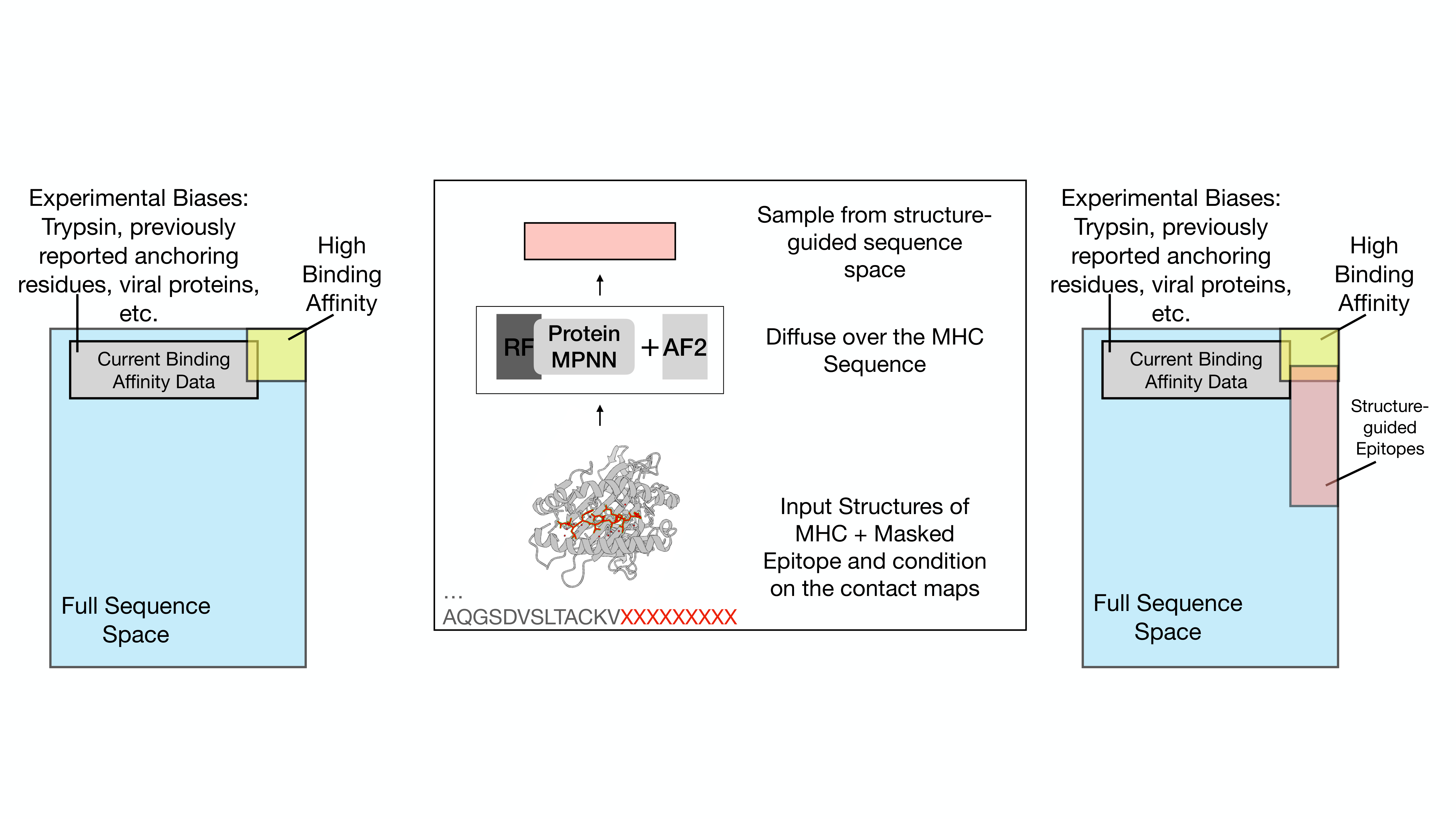
What if we could repurpose protein diffusion models as energy functions?
ProteinEBM does exactly that — turning a generative model into a scoring function
that can rank structures, predict conformational landscapes, and estimate mutation effects.
Protein Diffusion Models as Statistical Potentials
James P. Roney, Chenxi Ou, Sergey Ovchinnikov · bioRxiv, 2025
Alright, so here's the setup. Diffusion models have been crushing it in protein structure generation — you've probably heard of RFdiffusion and friends. But this paper asks a really clever question: what if instead of just using these models to generate new proteins, we could use them as energy functions? Like, what if the model's learned score function could tell us how "good" a given protein conformation is?
That's basically what ProteinEBM is. They take a pre-trained protein diffusion model and derive an energy-based model from it. The key insight is that the denoising process in a diffusion model implicitly learns a probability distribution over protein structures. If you can extract the energy landscape from that distribution, you've got yourself a statistical potential that knows what realistic protein conformations look like.
What's cool is how broadly useful this turns out to be. They show that ProteinEBM can rank the correctness of protein structures (which fold is more native-like?), do actual structure prediction from sequence, sample multiple conformational states of flexible proteins, and even predict the energetic effects of point mutations. And it's competitive with or better than both traditional physics-based force fields and other ML methods across these tasks.
The mutation effect prediction part is particularly interesting to me. There's been a ton of work on predicting delta-delta-G from sequence-based models, but having a structure-aware energy function that can do this feels like the right approach. You're literally asking "how does this mutation change the energy landscape?" which is what's actually happening physically.
The big picture here is that generative models contain way more information than we typically extract from them. We train them to generate, but the learned distribution itself is a rich model of protein physics. This paper is a nice reminder that sometimes the most interesting applications of a model aren't the ones it was originally trained for.
Read the paper
Designing Novel Solenoid Proteins with In Silico Evolution
Pretorius, Nikov, Washio, Florent, Taunt, Ovchinnikov, Murray · Communications Chemistry 2025
Solenoid proteins are nature's modular building blocks. This paper
uses AlphaFold2 as an oracle inside a genetic algorithm to design entirely
new solenoid folds — and 20% of them actually work in the lab.
Designing Novel Solenoid Proteins with In Silico Evolution
Daniella Pretorius, Georgi I. Nikov, Kono Washio, Steve-William Florent, Henry N. Taunt, Sergey Ovchinnikov, James W. Murray · Communications Chemistry (Nature), 2025
Solenoid proteins are one of those protein architectures that feel almost too elegant. They're built from repeating structural units — think of them like a spiral staircase made of protein. Nature uses them everywhere: from beta-helices in antifreeze proteins to TPR repeats that mediate protein-protein interactions. But the natural repertoire of solenoid topologies is limited. What if we could design new ones?
This paper takes a really fun approach. Instead of using the fancy generative models that everyone's been working on lately, they go old school: a genetic algorithm. But with a twist — they use AlphaFold2 as the fitness function. So you have a population of sequences that evolve over generations, and at each step, AlphaFold2 tells them which sequences fold into the desired solenoid topology. There's also a solenoid discrimination network that helps filter candidates.
What I like about this is the simplicity of the concept. You don't need to train a specialized generative model. You just need a good oracle (AlphaFold2) and a search strategy (the genetic algorithm). It's kind of like directed evolution, but entirely in silico. And it works — they designed alpha-solenoids, beta-solenoids, and alpha-beta solenoids, spanning both natural topologies and folds that haven't been seen in nature.
The experimental validation is solid too. They picked a handful of designs, expressed them in E. coli, and got five that were highly stable. That's a 20% experimental success rate, which is actually pretty good for de novo protein design. It's not 100%, but for a method that's essentially guessing-and-checking with AlphaFold2, it's encouraging.
One thing that struck me is how this approach reveals that the space of possible solenoid topologies is larger than what nature has explored. There are stable folds out there that evolution just never stumbled upon. That's a pretty cool thought — we're not just redesigning nature, we're expanding the protein universe.
Read the paper
CIRPIN: Learning Circular Permutation-Invariant Representations to Uncover Putative Protein Homologs
Kolodziej, Abulnaga, Ovchinnikov · bioRxiv 2025
Most structure comparison tools miss proteins that are related by circular permutation.
CIRPIN fixes this with a clever graph neural network that doesn't care where the chain starts —
uncovering thousands of hidden evolutionary relationships.
CIRPIN: Learning Circular Permutation-Invariant Representations
Aiden R. Kolodziej, S. Mazdak Abulnaga, Sergey Ovchinnikov · bioRxiv, 2025
Here's a problem that's been hiding in plain sight for years. Say you have two proteins that fold into essentially the same 3D shape, but one of them has its chain "rewired" — the N-terminus of one lines up with some internal position of the other. This is called circular permutation, and it's surprisingly common in nature. Proteins can evolve to swap where their chain starts and ends while keeping the same overall fold.
The problem? Most of our structural comparison tools — things like TM-align, Foldseek, DALI — are sensitive to chain topology. They compare structures in a way that cares about which residue comes first, second, third in the linear sequence. So two circularly permuted proteins that look basically identical in 3D can score as completely unrelated. That's a pretty big blind spot.
CIRPIN tackles this head-on with a graph neural network that's explicitly designed to be invariant to circular permutations. The clever bit is the data augmentation: during training, they synthetically circularly permute proteins so the model learns representations that are the same regardless of where you cut the chain. It's one of those ideas that seems obvious in hindsight but actually requires careful implementation.
The results are striking. They search through the SCOPe database and the AlphaFold Cluster Representatives and find thousands of novel circularly permuted pairs that previous methods completely missed. My favorite finding is that PDZ domains — one of the most studied protein families in structural biology — naturally exist in four distinct circularly permuted forms. Four! And we didn't know about some of these until CIRPIN found them.
This matters because circular permutation isn't just a structural curiosity. It can change a protein's function, stability, and interactions. If our homology detection tools are systematically missing these relationships, we're leaving a lot of biology on the table. CIRPIN fills that gap nicely.
Read the paper
Hit or Miss: Understanding Emergence and Absence of Homo-oligomeric Contacts in Protein Language Models
Zhang, Akiyama, Cho, Jajoo, Ovchinnikov · bioRxiv 2025
Protein language models are trained on single chains, yet they somehow learn about
protein-protein interfaces. This paper digs into how and why — and finds that
bigger models keep getting better at inter-chain contacts even after intra-chain accuracy plateaus.
Hit or Miss: Understanding Emergence and Absence of Homo-oligomeric Contacts in Protein Language Models
Zhidian Zhang, Yo Akiyama, Yehlin Cho, Samarth Jajoo, Sergey Ovchinnikov · bioRxiv, 2025
This one scratches a theoretical itch I've had for a while. Protein language models like ESM-2 are trained on individual protein sequences — single chains, one at a time. They never see protein complexes during training. And yet, when you extract attention maps or contact predictions from these models, they somehow know about inter-subunit contacts in homo-oligomers (proteins that assemble into copies of themselves). How?
The authors dig into this with a really systematic analysis. First, the scaling behavior is fascinating: as you make the model bigger, its ability to predict inter-chain contacts keeps improving, even after single-chain contact prediction accuracy has plateaued. That's a striking emergent property — the model discovers quaternary structure information as a byproduct of learning more about individual sequences.
There's also a cool comparison between ESM-2 and MSA-based approaches (MSA Pairformer). When you restrict multiple sequence alignments to close evolutionary neighbors, the MSA approach edges out ESM-2 on interface contact recovery (0.44 vs 0.33). But ESM-2 doesn't need any alignment at all — it works from a single sequence. So there's a tradeoff between the coevolutionary signal you get from alignments and the implicit knowledge baked into a massive language model.
One really practical result: the largest ESM-2 model can accurately distinguish genuine biological interfaces from crystal packing artifacts. This is a classic problem in structural biology — when you solve a crystal structure, you see contacts between protein copies in the crystal lattice, and it's not always obvious which of those contacts are biologically real. Having a sequence-based model that can make this call is genuinely useful.
The "hit or miss" framing in the title is apt. The model doesn't learn all interfaces equally. Some it nails, others it completely misses. Understanding which ones and why is where the real mechanistic insight lies, and this paper makes good progress on that front.
Read the paper
Assessing the Utility of Coevolution-Based Residue–Residue Contact Predictions in a Sequence- and Structure-Rich Era
Kamisetty, Ovchinnikov, Baker · PNAS 2013
The 2013 paper that helped establish when coevolution-based contact
prediction is actually useful. A foundational work that set the stage for
everything from direct coupling analysis to AlphaFold.
Assessing the Utility of Coevolution-Based Contact Predictions
Hetunandan Kamisetty, Sergey Ovchinnikov, David Baker · PNAS, 2013
Going back to 2013 for this one, and honestly, reading it now with the benefit of hindsight is wild. This is from the era before AlphaFold, before protein language models, before any of the deep learning revolution in structural biology. The big question was: can we use the patterns of correlated mutations in multiple sequence alignments to predict which residues are in physical contact in a protein's 3D structure?
The answer had been "sort of, sometimes" for a while. What this paper does is lay out clearly when coevolution-based contact prediction actually works well and when it doesn't. The key finding is a simple but powerful rule of thumb: you need at least 5L non-redundant aligned sequences (where L is the protein length) for the predictions to be reliable. Below that threshold, the signal-to-noise ratio just isn't good enough.
They also make a practical argument about when accurate contacts are most useful for structure modeling. If you already have a close homolog with a known structure, the contacts don't add much — you can just thread your sequence onto the known template. But when your best structural template is distant, predicted contacts can bridge the gap and dramatically improve models. It's about finding the sweet spot where you have enough sequences for coevolution but not enough structural information from homology alone.
What makes this paper historically important is that it helped establish the credibility of coevolution-based methods at a time when many people were skeptical. The computational community needed convincing that these predictions were accurate enough to be useful, and this paper provided the benchmarks and the practical guidelines that moved the field forward.
Looking back, this work was laying the intellectual groundwork for everything that followed. The idea that evolutionary information contains structural information — that patterns in sequences encode 3D contacts — is the same insight that powers AlphaFold's Evoformer and the MSA processing in ESMFold. The methods got fancier, but the core principle established here still holds.
Read the paper
More papers I find interesting
De Novo Design of Protein Structure and Function with RFdiffusion
Watson, Juergens, Bennett et al. · Nature 2023
The paper that brought diffusion models to protein design in a big way.
RFdiffusion generates protein backbones from scratch and can design binders,
symmetric assemblies, and enzyme scaffolds — many validated experimentally.
De Novo Design of Protein Structure and Function with RFdiffusion
Joseph L. Watson, David Juergens, Nathaniel R. Bennett et al. · Nature, 2023
If there's one paper that made everyone in structural biology sit up and pay attention to diffusion models, it's this one. RFdiffusion takes the RoseTTAFold structure prediction architecture and fine-tunes it on a denoising task: starting from random noise, iteratively denoise until you get a plausible protein backbone. It's the same idea as image diffusion models (Stable Diffusion, DALL-E), but for 3D protein structures.
What makes this paper really stand out isn't just the method — it's the sheer range of design challenges they tackle. Unconditional monomer design? Check. Topology-constrained design? Check. Protein binder design where you specify a target surface and the model hallucates a complementary protein? Check. Symmetric homo-oligomer design? Also check. Enzyme active site scaffolding? Yep, that too.
And they don't just design things computationally. There's extensive experimental validation. They express hundreds of designs, characterize them with various biophysical methods, and even solve cryo-EM structures of some symmetric assemblies. The designed binders actually bind. The symmetric assemblies actually assemble. It's not just pretty pictures from a generative model — these things work in the real world.
The underlying trick is elegant: by training on the denoising objective, the model learns a gradient field over protein structure space that points "uphill" toward realistic folds. Conditioning the denoising on different constraints (target surface, symmetry operations, motif coordinates) lets you steer generation toward specific design goals. It's a really flexible framework.
For my own work on pMHC design, RFdiffusion is the backbone generation engine. So I have a personal stake in understanding this paper well. The conditioning mechanism is what lets us specify MHC groove geometry and get peptide backbones that fit — it's a direct application of the tools developed here.
Read the paper
Evolutionary-Scale Prediction of Atomic-Level Protein Structure with a Language Model (ESMFold)
Lin, Abanades, Rao, Johnson, Rives et al. · Science 2023
What if you could predict protein structure from a single sequence, no alignment needed?
ESMFold does this at AlphaFold-like accuracy with a 15 billion parameter language model,
enabling structure prediction for 600+ million metagenomic proteins.
ESMFold: Protein Structure from a Language Model
Zeming Lin, Halil Akin, Roshan Rao, Jeff Johnson, Alexander Rives et al. · Science, 2023
AlphaFold2 was a revolution, but it has a practical bottleneck: it needs multiple sequence alignments (MSAs). For every protein you want to fold, you first have to search databases, build an alignment, and then run the model. This is slow, and for orphan proteins with few homologs, the alignments can be thin and noisy.
ESMFold's pitch is simple: what if a large enough language model, trained on enough protein sequences, could learn the evolutionary information implicitly? No alignment needed — just give it a single sequence and it predicts the structure. The language model (ESM-2, up to 15 billion parameters) acts as a replacement for the entire MSA pipeline.
The accuracy is impressive. It doesn't quite match AlphaFold2 on every benchmark, but it gets close enough to be extremely useful, especially because it's orders of magnitude faster. And for proteins where building a good MSA is hard or impossible (think metagenomic sequences with no close relatives), ESMFold can still make predictions. That's a huge practical advantage.
The real flex in this paper is the ESM Metagenomic Atlas. They ran ESMFold on 617 million metagenomic protein sequences and predicted structures for all of them. This was simply not feasible with AlphaFold2's MSA requirement. The resulting atlas reveals an enormous amount of structural diversity in environmental sequences — folds that we've never seen in lab-characterized proteins.
For the protein language model community, ESMFold is an important proof of concept. It shows that the representations learned by masked language modeling on protein sequences contain enough structural information to fold proteins. The model isn't just learning sequence patterns — it's learning physics. That's conceptually very satisfying and has big implications for how we think about what these models are actually capturing.
Read the paper
Simulating 500 Million Years of Evolution with a Language Model (ESM3)
Hayes, Rao, Akin et al. · Science 2025
ESM3 is a 98-billion-parameter multimodal model that reasons over protein sequence,
structure, and function simultaneously. It designed a novel fluorescent protein with
only 58% identity to anything in nature — equivalent to 500 million years of evolution.
Simulating 500 Million Years of Evolution with ESM3
Thomas Hayes, Roshan Rao, Halil Akin et al. · Science, 2025
Okay, so this paper is kind of a big deal. ESM3 is a 98-billion-parameter model that doesn't just look at protein sequences — it simultaneously reasons over sequence, structure, and function. It's multimodal in the protein sense: trained on 3.15 billion sequences, 236 million structures, and 539 million function annotations. That's 771 billion tokens total.
The headline result is esmGFP, a computationally designed fluorescent protein that has only 58% sequence identity to any known fluorescent protein. To put that in perspective, natural GFP variants across all of biology (jellyfish, corals, sea anemones) typically share more than 58% identity with each other. The authors estimate this level of divergence would take over 500 million years of natural evolution to achieve. And yet, when they expressed esmGFP in the lab, it fluoresced. It actually worked.
That result is remarkable, but let's be honest about what it means and doesn't mean. The model didn't "simulate evolution" in any mechanistic sense — it's not running a phylogenetic process. What it did was learn the manifold of viable protein sequences well enough to navigate far from known sequences while staying on the functional surface. That's impressive in a different way. It means the model has a deep enough understanding of protein fitness landscapes to make large creative leaps.
The multimodal training is key here. By jointly learning from sequence, structure, and function data, ESM3 can be prompted in any combination of these modalities. Want a protein with a specific fold? Prompt with structure. Want a specific function? Prompt with function tokens. Want both? Prompt with both. This flexibility is what enables the GFP design — they could specify "be fluorescent" and "fold like a beta-barrel" and let the model figure out a sequence that satisfies both constraints.
The scale is also hard to ignore. 98 billion parameters is massive, and the training data spans essentially all known protein diversity. Whether this approach scales further (can you design even more novel proteins with even bigger models?) is an open question, but ESM3 suggests the answer is probably yes.
Read the paper
Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3
Abramson, Adler, Dunger et al. · Nature 2024
AlphaFold 3 moves beyond proteins to predict the structures of complexes involving
DNA, RNA, small molecules, and ions — with a diffusion-based architecture that
substantially outperforms specialized tools for drug-like interactions.
AlphaFold 3: Biomolecular Interactions, Not Just Proteins
Josh Abramson, Jonas Adler, Jack Dunger et al. · Nature, 2024
AlphaFold 2 was a protein structure prediction tool. AlphaFold 3 is something broader — it's a biomolecular interaction prediction tool. The difference matters. Biology doesn't run on isolated proteins; it runs on proteins bound to DNA, RNA, small molecules, metal ions, and each other. AF3 handles all of these in a unified framework.
The architectural change is significant. AF2 used an iterative recycling mechanism with specialized modules for MSA processing and pairwise reasoning. AF3 replaces much of this with a diffusion-based approach. The model predicts atomic coordinates directly through a denoising process, which turns out to be more natural for handling the diversity of molecular types (you can't really use the same residue-level representation for a protein and a small molecule).
The results on protein-ligand interactions are what caught the drug discovery community's attention. AF3 substantially outperforms established docking tools for predicting how drugs bind to proteins. That's a big deal because molecular docking has been a cornerstone of computational drug discovery for decades, and having a learned model surpass physics-based approaches suggests a real paradigm shift.
For protein-nucleic acid interactions, AF3 also beats specialized tools. And for antibody-antigen prediction — notoriously one of the hardest problems in structural biology because CDR loops are so flexible — it significantly improves over AF2-Multimer. These are all areas where having accurate structural predictions has immediate practical applications.
The catch (there's always a catch) is that diffusion-based structure prediction introduces stochasticity. You get different predictions on different runs, which is both a feature (you can sample conformational diversity) and a complication (you need to run multiple seeds and assess confidence carefully). But overall, AF3 represents a genuine step forward in our ability to model the full complexity of cellular molecular machinery.
Read the paper
Protein Language Models Learn Evolutionary Statistics of Interacting Sequence Motifs
Zhang, Wayment-Steele, Brixi, Wang, Kern, Ovchinnikov · PNAS 2024
What do protein language models actually learn? This paper shows ESM-2
stores coevolutionary statistics as motifs of pairwise contacts —
bridging the gap between classical coevolution and modern deep learning.
What Do Protein Language Models Actually Learn?
Zhidian Zhang, Hannah K. Wayment-Steele, Garyk Brixi, Haobo Wang, Dorothee Kern, Sergey Ovchinnikov · PNAS, 2024
This paper is important because it tries to open the black box. We all know that protein language models like ESM-2 learn useful representations — you can predict structure, function, fitness effects, and all sorts of things from their embeddings. But what exactly are they learning? Is it deep, emergent reasoning about protein physics? Or something simpler?
The answer, at least for contact prediction, turns out to be more interpretable than you might expect. The authors develop an unsupervised method to probe what ESM-2 has learned and find that the model stores statistics of coevolving residues — essentially the same coevolutionary signal that classical methods like direct coupling analysis (DCA) extract from multiple sequence alignments. But instead of computing these statistics from alignments at inference time, ESM-2 has memorized them during pre-training.
Even more specifically, ESM-2 predicts contacts by storing "motifs" — small groups of pairwise contacts that tend to co-occur. It's not doing some deep chain of reasoning across the entire protein. It's more like pattern matching against a library of local contact patterns. This is both reassuring (the model is learning something real and interpretable) and a little humbling (it's not doing anything as sophisticated as we might have imagined).
One surprising finding: ESM-2 doesn't actually need the full sequence context to predict inter-residue contacts. You can mask or truncate large portions of the sequence and the model still predicts contacts for the remaining region. This suggests the model is doing local pattern recognition rather than global inference, which is consistent with the motif-based explanation.
For the field, this kind of mechanistic understanding is crucial. If we know what the model learns, we can design better models, identify failure modes, and figure out what information is still missing. It also connects the protein language model revolution back to the coevolution-based methods that came before — showing that there's a continuous intellectual thread from DCA to transformer attention maps.
Read the paper
Last updated: March 9, 2026
Goal: Train a transformer encoder from scratch on CATH 4.2 dataset (18k proteins) to jointly predict CATH class/architecture labels AND inter-residue contact maps from sequence alone. The learned embeddings encode spatial proximity information needed for Stage 2.
Architecture
ContactClassifier — 1.2M params, dim=128, 2 transformer towers, d_pair=64, 2 contact prediction blocks with outer product mean
Training
Single GPU (1080 Ti), batch_size=24, lr=2e-4 with warmup cosine schedule, patience=15 early stopping
Resilience
Per-epoch checkpoints with auto-resume, CSV loss logging, self-resubmitting watchdog system on SLURM
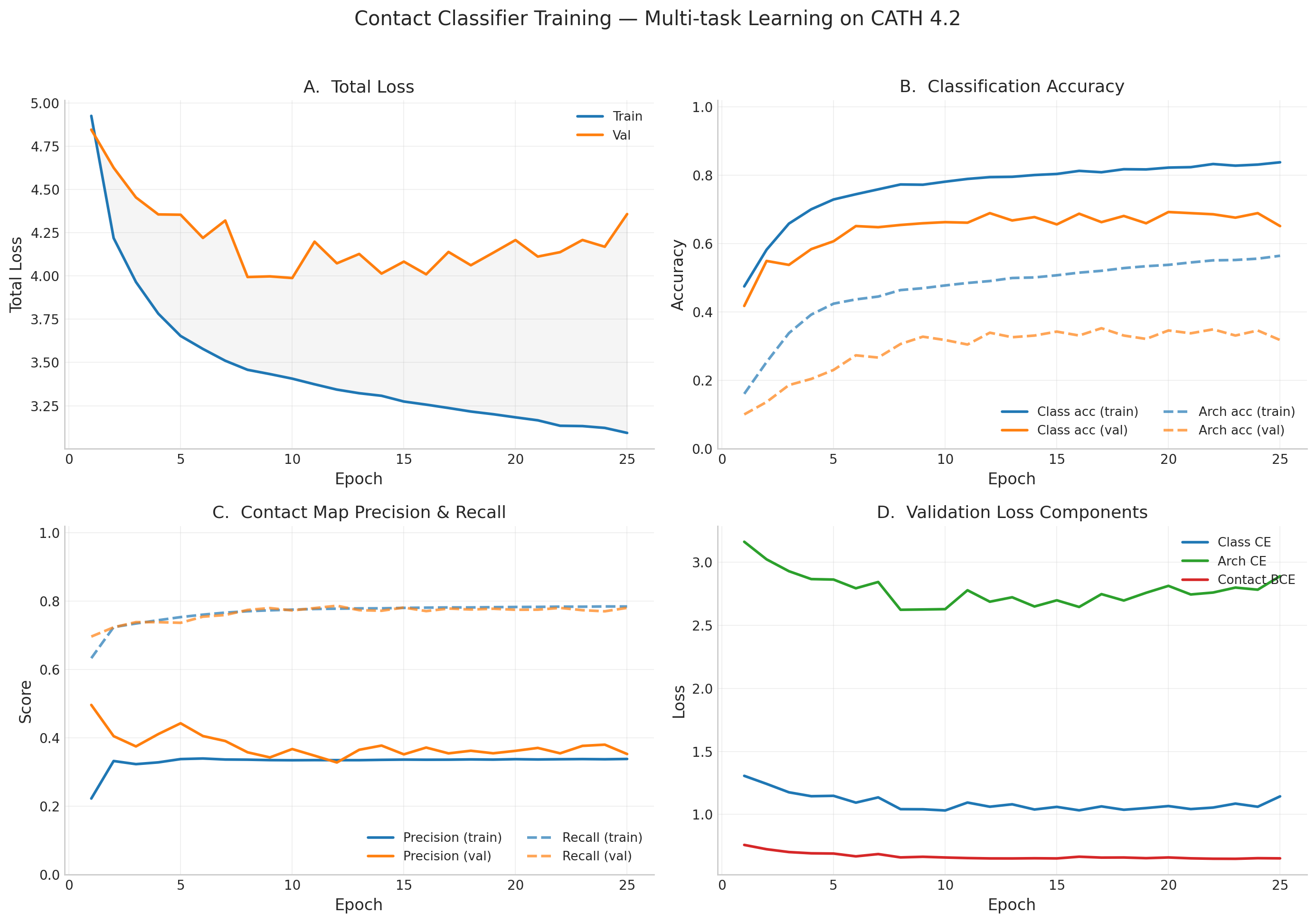
Training Progress (all 25 epochs — early stopped)
| Epoch |
Val Total Loss |
Train Class Acc |
Val Class Acc |
Train Arch Acc |
Contact Recall (Val) |
Contact BCE (Val) |
LR |
Status |
| 1 |
4.846 |
47.5% |
41.8% |
16.1% |
69.6% |
0.759 |
3.33e-05 |
NEW BEST |
| 2 |
4.626 |
58.2% |
54.9% |
25.4% |
72.3% |
0.725 |
6.67e-05 |
NEW BEST |
| 3 |
4.454 |
65.9% |
53.8% |
33.8% |
73.9% |
0.702 |
1.00e-04 |
NEW BEST |
| 4 |
4.356 |
70.0% |
58.4% |
39.2% |
73.8% |
0.693 |
1.33e-04 |
NEW BEST |
| 5 |
4.354 |
72.9% |
60.7% |
42.4% |
73.7% |
0.691 |
1.67e-04 |
NEW BEST |
| 6 |
4.220 |
74.4% |
65.1% |
43.7% |
75.5% |
0.668 |
2.00e-04 |
NEW BEST |
| 7 |
4.321 |
75.9% |
64.8% |
44.5% |
75.9% |
0.686 |
2.00e-04 |
pat 1 |
| 8 |
3.994 |
77.3% |
65.5% |
46.4% |
77.4% |
0.660 |
1.99e-04 |
NEW BEST |
| 9 |
3.998 |
77.2% |
66.0% |
47.0% |
78.0% |
0.665 |
1.98e-04 |
pat 1 |
| 10 |
3.988 |
78.1% |
66.3% |
47.8% |
77.3% |
0.659 |
1.97e-04 |
BEST (final) |
| 11 |
4.199 |
78.9% |
66.1% |
48.5% |
78.0% |
0.655 |
1.96e-04 |
pat 1 |
| 12 |
4.073 |
79.4% |
68.9% |
49.1% |
78.7% |
0.652 |
1.94e-04 |
pat 2 |
| 13 |
4.128 |
79.5% |
66.8% |
49.9% |
77.4% |
0.652 |
1.92e-04 |
pat 3 |
| 14 |
4.014 |
80.1% |
67.8% |
50.1% |
77.2% |
0.653 |
1.89e-04 |
pat 4 |
| 15 |
4.083 |
80.4% |
65.6% |
50.7% |
78.1% |
0.652 |
1.87e-04 |
pat 5 |
| 16 |
4.010 |
81.3% |
68.8% |
51.5% |
77.1% |
0.666 |
1.84e-04 |
pat 6 |
| 17 |
4.139 |
80.9% |
66.3% |
52.1% |
77.8% |
0.659 |
1.80e-04 |
pat 7 |
| 18 |
4.062 |
81.8% |
68.1% |
52.8% |
77.5% |
0.659 |
1.77e-04 |
pat 8 |
| 19 |
4.134 |
81.7% |
66.0% |
53.4% |
77.8% |
0.654 |
1.73e-04 |
pat 9 |
| 20 |
4.208 |
82.2% |
69.2% |
53.8% |
77.5% |
0.660 |
1.69e-04 |
pat 10 |
| 21 |
4.112 |
82.4% |
68.9% |
54.5% |
77.5% |
0.653 |
1.64e-04 |
pat 11 |
| 22 |
4.138 |
83.3% |
68.6% |
55.1% |
78.0% |
0.650 |
1.60e-04 |
pat 12 |
| 23 |
4.208 |
82.8% |
67.6% |
55.2% |
77.3% |
0.649 |
1.55e-04 |
pat 13 |
| 24 |
4.169 |
83.1% |
68.9% |
55.6% |
77.0% |
0.654 |
1.50e-04 |
pat 14 |
| 25 |
4.358 |
83.8% |
65.1% |
56.4% |
78.0% |
0.652 |
1.45e-04 |
EARLY STOP |
Final Results
- Best val loss: 3.988 at epoch 10 (best weights saved)
- Val class accuracy: 66.3% (4-way CATH class), Val architecture accuracy: 31.8% (38+ architectures)
- Contact recall: 77.3%, Contact BCE: 0.659 — model successfully learned spatial proximity from sequence
- Train class accuracy: 78.1%, Train arch accuracy: 47.8%
- Early stopped at epoch 25 (patience 15) — val loss plateaued after epoch 10
- 1.2M params, trained from scratch on CATH 4.2 (18k proteins)
- Training survived 7 SLURM job allocations with checkpoint resume
Training Curves
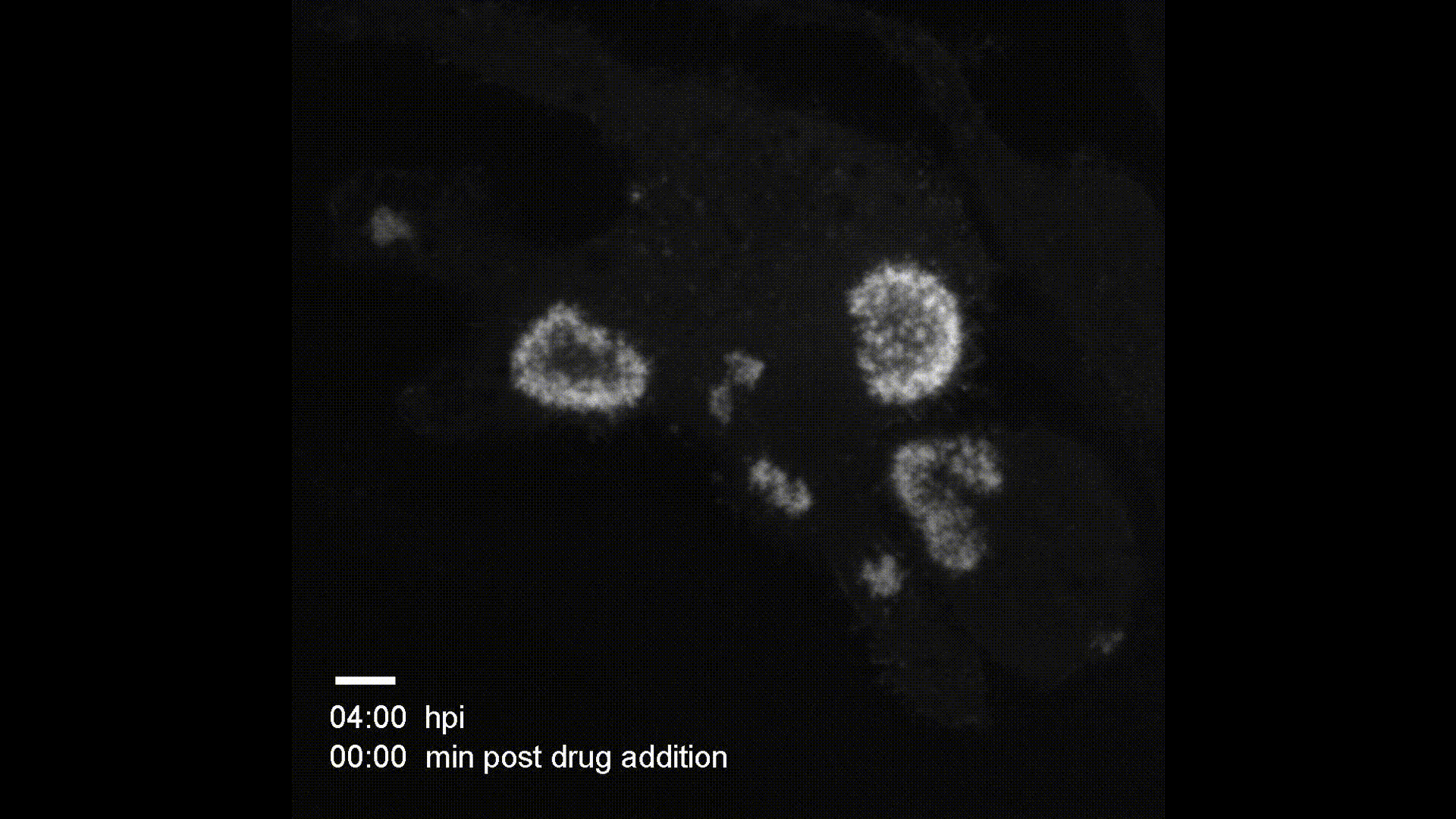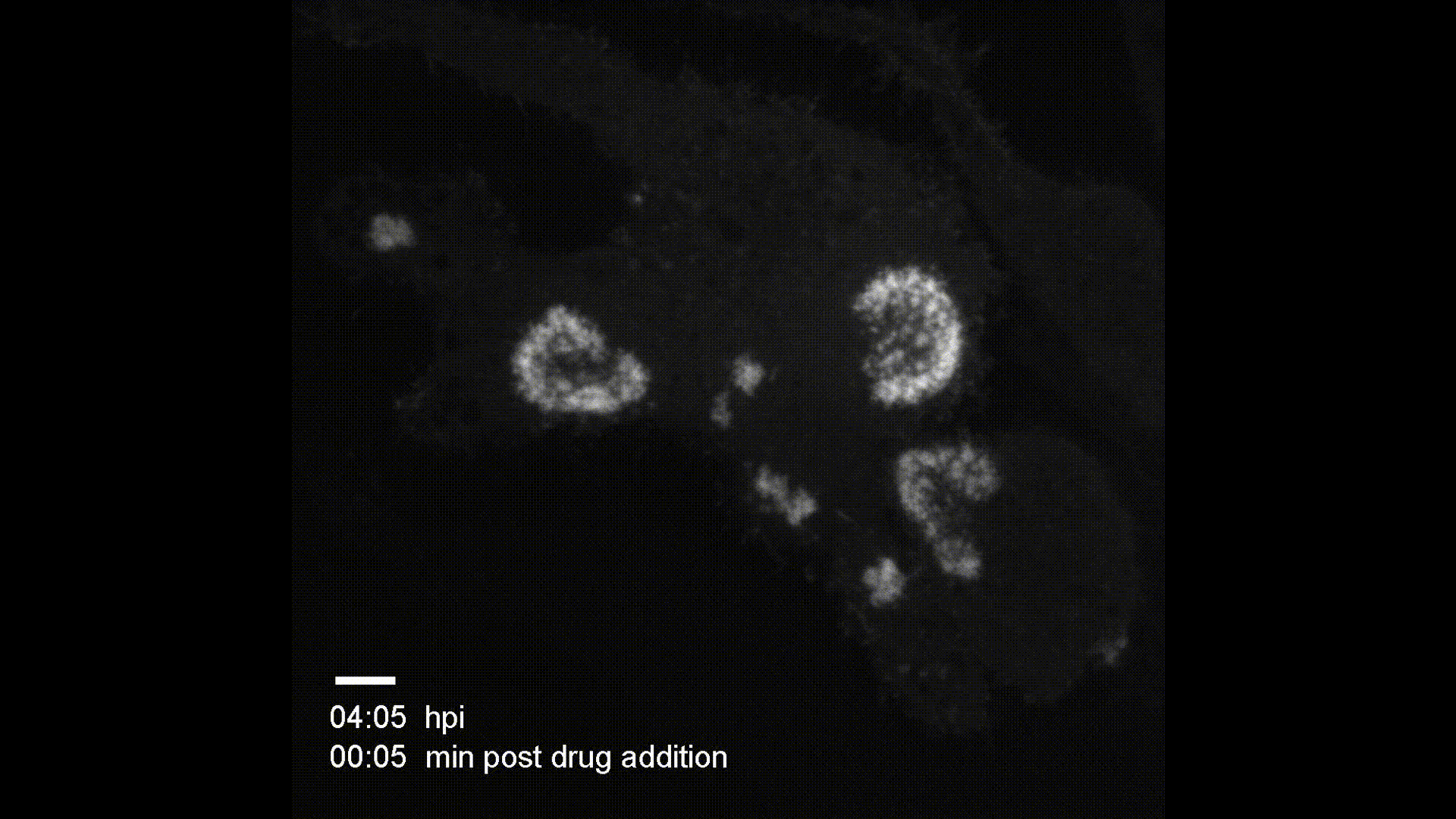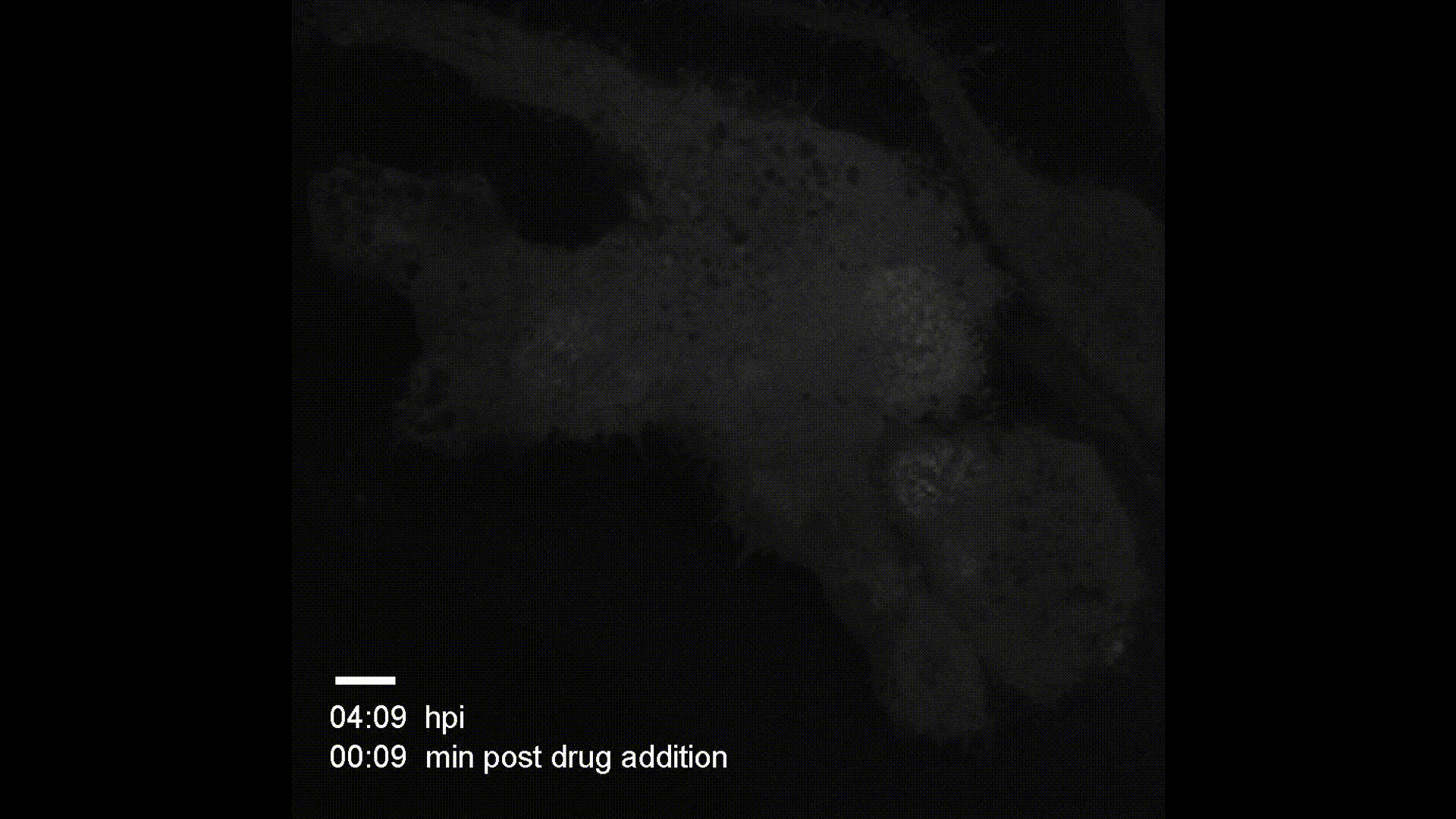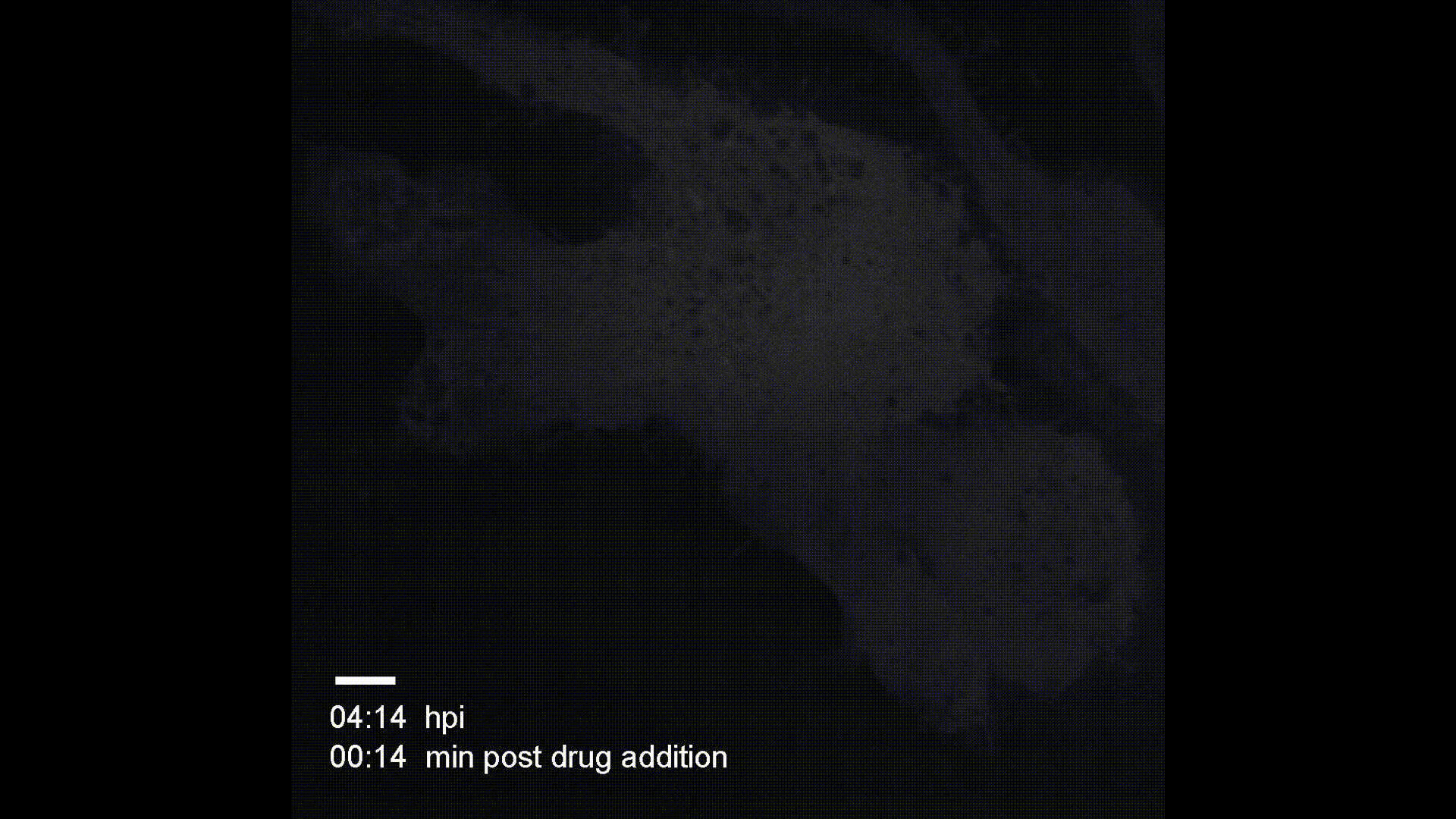
Contact Map Predictions (3 test proteins)
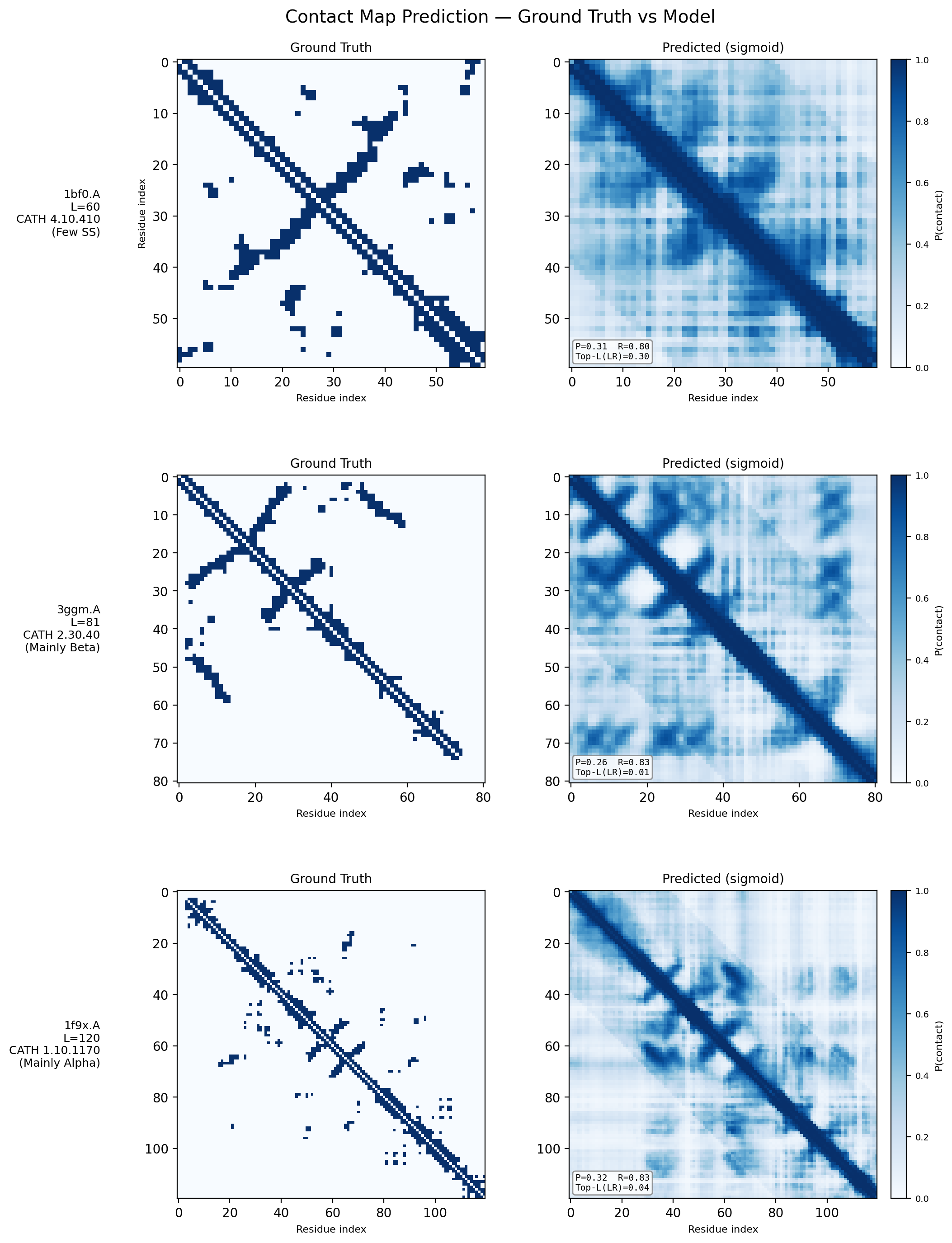
Each row shows a held-out test protein from a different CATH structural class. The left column is the ground truth contact map (binary: two Cα atoms < 8Å apart), and the right column is the model’s predicted probability of contact from sequence alone. Metrics (precision P, recall R, and Top-L long-range accuracy) are annotated on each prediction panel.
- 1bf0.A (L=60, Few Secondary Structure): A small protein with sparse, irregular contacts. The model captures the overall topology despite limited structural regularity.
- 3ggm.A (L=81, Mainly Beta): Beta-sheet proteins produce characteristic off-diagonal block patterns from strand–strand hydrogen bonding. The model recovers these long-range parallel and anti-parallel strand pairings well.
- 1f9x.A (L=120, Mainly Alpha): Alpha-helical proteins show strong banded diagonal patterns from helix-internal i→i+4 contacts. The model reproduces both the local helical periodicity and inter-helix contacts at larger separations.
These results demonstrate that a 1.2M-parameter transformer encoder trained from scratch on CATH 4.2 (~18k proteins) can learn meaningful spatial proximity signals across all major fold classes — without any pretrained language model or evolutionary information.
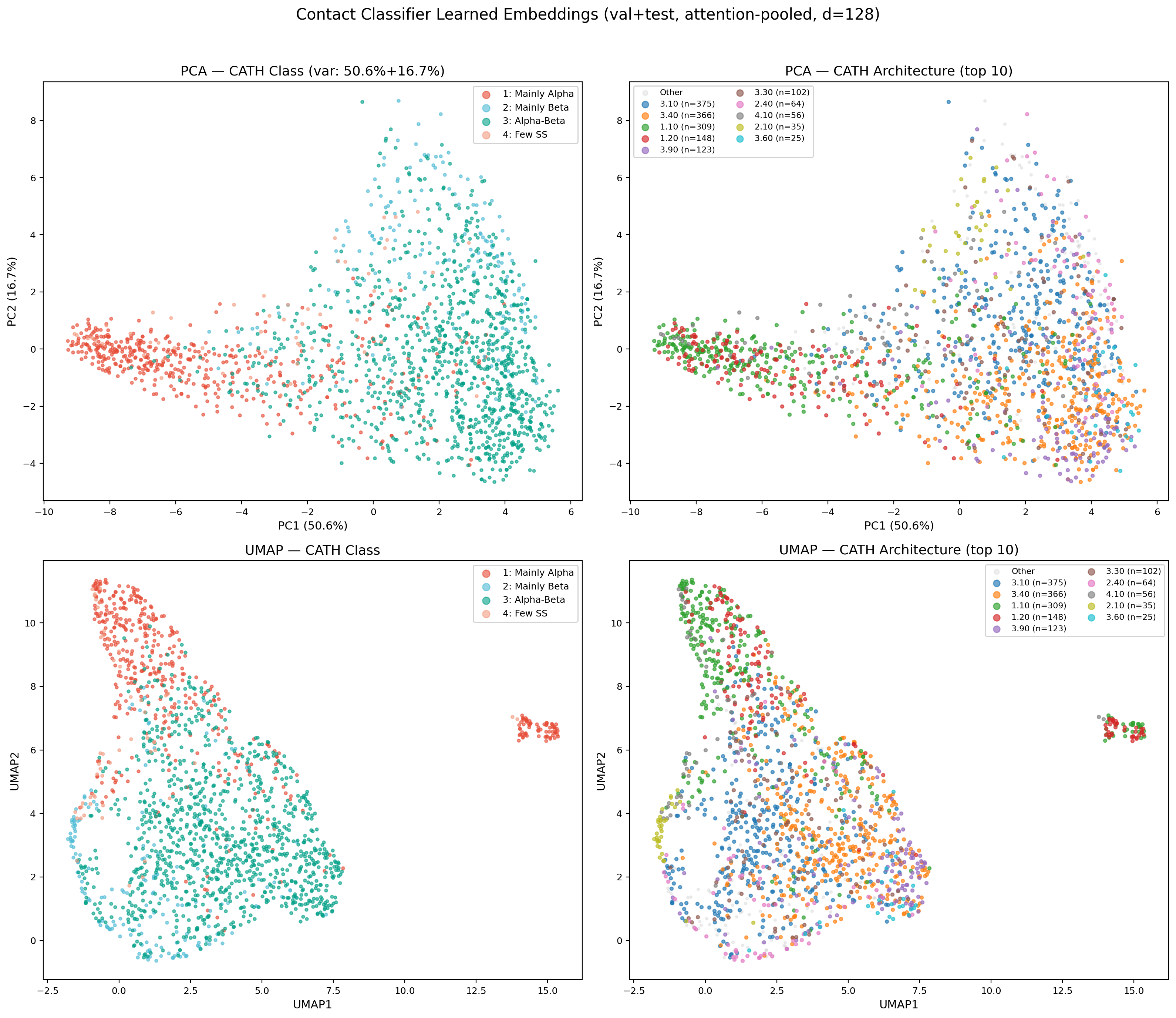
Learned Embedding Space (PCA & UMAP)
Attention-pooled protein embeddings (128-dim) from the encoder’s val+test set, projected via PCA and UMAP. The encoder learns to separate CATH classes without explicit contrastive loss — mainly-alpha and mainly-beta proteins form distinct clusters, while alpha-beta proteins span the intermediate region. UMAP reveals finer sub-structure at the architecture level, with several CATH architectures forming tight, well-separated clusters (e.g., 3.40 Rossmann fold, 1.10 orthogonal bundle). This confirms the multi-task training objective (classification + contact prediction) produces structurally meaningful representations suitable for conditioning the downstream diffusion model.
Goal: Generate realistic protein backbone structures (Cα coordinates) conditioned on sequence, using contact-aware embeddings from Stage 1. v12 scales IPA capacity 4.1x over v11b (31.1M params) to resolve the gradient competition collapse that limited v11b — same validated loss design, wider single representation, deeper frame update MLP.
Architecture
IPA denoiser (8 layers, 8 heads, 8 query points) + independent aux pair stack (64-dim) + frozen ContactClassifier encoder — 31.1M params (29.9M trainable, 1.2M frozen); d_ipa_hidden=512, d_ipa_ffn=1024, 2-layer FrameUpdate MLP
Training
Single RTX 2080 Ti (11.5 GB), batch_size=4, grad_accum=4 (eff=16), LR=2e-5, gradient checkpointing, T=1000, DDIM-50 eval, SC=0.25, w_frame_rot=0.5
Dataset
CATH 4.2: 18,024 train / 608 val proteins, max 125 residues
Why IPA? The v10 Ceiling
v10 used an 8-layer EGNN denoiser that learned pairwise distance statistics well (dist_mse 60% below random)
but could not learn protein topology. FAPE stayed at its random baseline (~1.31) across 21 epochs and
TM-score peaked at 0.131 (random ~0.10). EGNN has no concept of local reference frames — it reasons about
distances, not backbone geometry. IPA solves this by maintaining and refining per-residue rigid-body frames
(rotation + translation) through 3D point attention in local coordinate systems.
v11b Key Changes (from v11)
- Frame rotation loss (w=0.5) — direct angular distance:
1 - cos(angle) between learned R and Gram-Schmidt ground truth. Gives rotation quaternions direct gradient for the first time.
- FAPE with learned frames —
fape_loss_with_frames uses R_pred from IPA layers instead of rebuilding from coordinates. The R → FAPE gradient path is now intact.
- LR halved — 2e-5 (from 5e-5) to prevent overshooting now that rotations receive gradient
- Self-conditioning reduced — SC probability 0.25 (from 0.5) to let the model learn from scratch more
- Initialized from v11 E10 best — EMA weights only, fresh optimizer + scheduler
v11 Architecture (shared with v11b)
- IPA denoiser — each block: invariant point attention (scalar + 3D point Q/K/V in local frames + pair bias) → transition MLP → frame update (quaternion + translation, identity-biased init, composed in local frame for SE(3) equivariance)
- Fixed-scale coordinates — divide by 10Å instead of per-protein Rg normalization
- SNR-gated frame initialization — Gram-Schmidt frames from Cα at low noise, slerp to identity at high noise
- Independent aux pair stack (64-dim, 2 layers) with ordinal regression (32 bins)
v11b Loss Weights
| Loss |
v11 |
v11b |
Rationale |
| FAPE (w/ learned R) | 1.0 | 1.0 | v11b uses learned R_pred instead of Gram-Schmidt rebuilt frames |
| Frame Rot | — | 0.5 | NEW: direct angular loss on learned R vs ground truth. The v11b fix. |
| Bond | 3.0 | 3.0 | — |
| Clash | 0.1 | 0.1 | — |
| Aux dist | 0.03 | 0.03 | — |
| Dist MSE | 1.0 | 1.0 | — |
| Chirality | 0.1 | 0.1 | — |
| Angle | 0.5 | 0.5 | — |
| Rg | 0.5 | 0.5 | — |
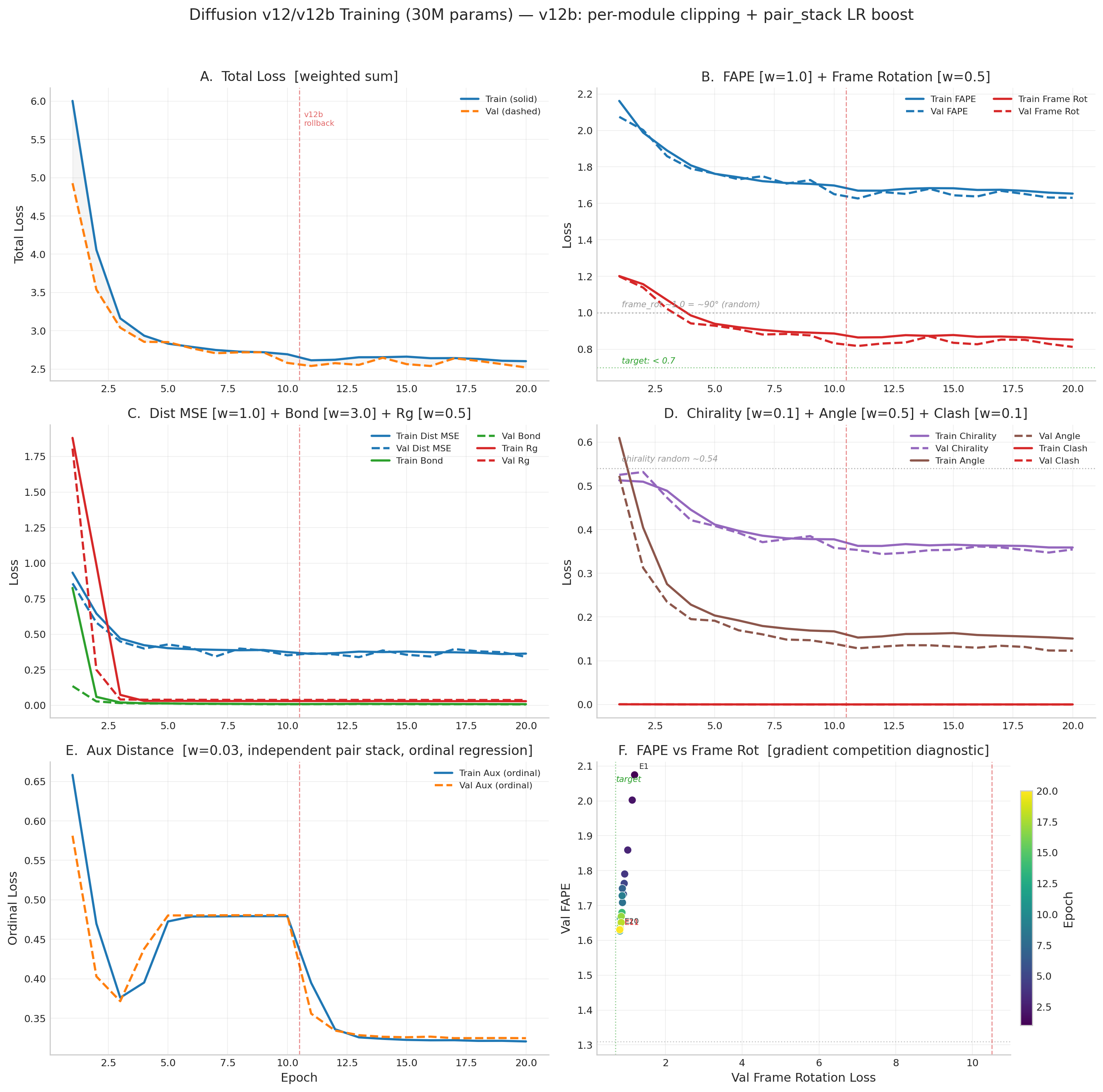
v12 Loss Curves (through Epoch 13)
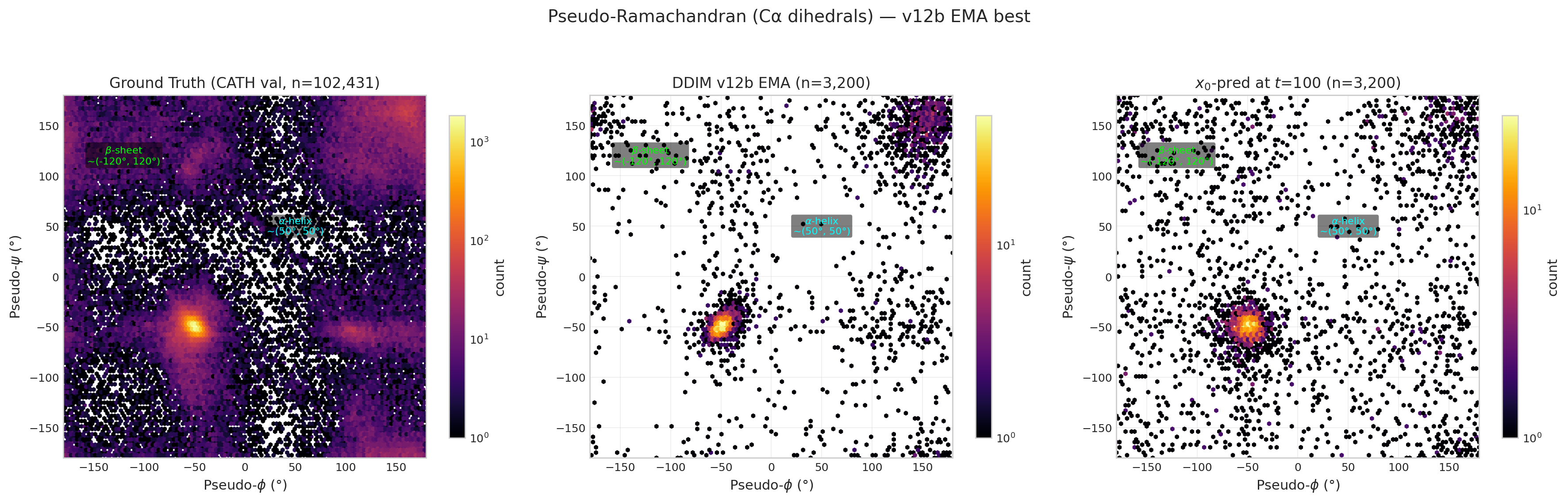
Pseudo-Ramachandran Analysis
Pseudo-dihedrals computed from consecutive Cα positions. Ground truth (left) shows clear α-helix (~50°,50°) and β-sheet (~−120°,120°) clusters. DDIM samples and low-noise x0_pred panels pending GPU evaluation.
v12 Training Progress
| Epoch |
Val Total |
Val FAPE |
Val Frame Rot |
Val Dist |
Val Bond |
DDIM TM |
DDIM RMSD |
Status |
| 1 |
4.928 |
2.074 |
1.199 |
0.857 |
0.135 |
0.101 |
15.28Å |
NEW BEST |
| 2 |
3.538 |
2.002 |
1.138 |
0.581 |
0.028 |
0.132 |
13.73Å |
NEW BEST |
| 3 |
3.041 |
1.859 |
1.022 |
0.449 |
0.016 |
0.118 |
15.33Å |
NEW BEST |
| 4 |
2.856 |
1.790 |
0.942 |
0.399 |
0.013 |
0.103 |
16.56Å |
NEW BEST |
| 5 |
2.852 |
1.763 |
0.929 |
0.428 |
0.013 |
0.101 |
16.88Å |
NEW BEST |
| 6 |
2.771 |
1.732 |
0.910 |
0.403 |
0.010 |
0.102 |
16.99Å |
NEW BEST |
| 7 |
2.707 |
1.749 |
0.881 |
0.343 |
0.010 |
0.099 |
17.26Å |
NEW BEST |
| 8 |
2.719 |
1.708 |
0.884 |
0.399 |
0.009 |
0.099 |
17.23Å |
pat 1 |
| 9 |
2.720 |
1.728 |
0.876 |
0.386 |
0.008 |
0.101 |
17.04Å |
pat 2 |
| 10 |
2.581 |
1.651 |
0.833 |
0.352 |
0.008 |
0.102 |
17.09Å |
NEW BEST |
| 11 |
2.685 |
1.684 |
0.864 |
0.391 |
0.009 |
0.098 |
17.41Å |
pat 1 |
| 12 |
2.831 |
1.804 |
0.905 |
0.391 |
0.009 |
0.087 |
18.46Å |
pat 2 |
| 13 |
2.822 |
1.814 |
0.913 |
0.370 |
0.009 |
0.078 |
19.54Å |
pat 3 |
|
— v12b rollback to E10 EMA — per-module grad clipping, pair_stack 3x LR, tripwire
|
| b1 |
2.541 |
1.627 |
0.818 |
0.366 |
0.008 |
0.102 |
16.98Å |
NEW BEST |
| b2 |
2.577 |
1.662 |
0.831 |
0.358 |
0.008 |
0.105 |
16.89Å |
pat 1 |
| b3 |
2.555 |
1.652 |
0.837 |
0.339 |
0.009 |
0.103 |
17.09Å |
pat 2 |
| b4 |
2.648 |
1.680 |
0.870 |
0.386 |
0.009 |
0.107 |
16.88Å |
pat 3 |
| b5 |
2.565 |
1.644 |
0.835 |
0.356 |
0.008 |
0.104 |
17.02Å |
pat 4 |
| b6 |
2.540 |
1.638 |
0.827 |
0.343 |
0.007 |
0.108 |
16.80Å |
pat 5 |
| b7 |
2.641 |
1.668 |
0.852 |
0.396 |
0.008 |
0.113 |
16.47Å |
pat 6 |
| b8 |
2.607 |
1.651 |
0.851 |
0.379 |
0.008 |
0.113 |
16.39Å |
pat 7 |
| b9 |
2.564 |
1.632 |
0.829 |
0.374 |
0.007 |
0.112 |
16.40Å |
pat 8 |
| b10 |
2.521 |
1.630 |
0.813 |
0.341 |
0.006 |
0.113 |
16.42Å |
NEW BEST |
| b11 |
2.553 |
1.633 |
0.829 |
0.362 |
0.006 |
0.110 |
16.52Å |
pat 1 |
| b12 |
2.558 |
1.628 |
0.835 |
0.370 |
0.007 |
0.111 |
16.56Å |
pat 2 |
| b13 |
2.534 |
1.635 |
0.826 |
0.343 |
0.007 |
0.110 |
16.64Å |
pat 3 |
| b14 |
2.500 |
1.606 |
0.814 |
0.349 |
0.006 |
0.107 |
17.13Å |
NEW BEST |
| b15 |
2.480 |
1.586 |
0.807 |
0.391 |
0.006 |
0.112 |
16.62Å |
NEW BEST |
| b16 |
2.469 |
1.564 |
0.780 |
0.347 |
0.006 |
0.112 |
16.62Å |
NEW BEST |
| b17 |
2.506 |
1.608 |
0.815 |
0.355 |
0.006 |
0.110 |
16.88Å |
pat 1 |
| b18 |
2.497 |
1.626 |
0.809 |
0.335 |
0.005 |
0.108 |
16.83Å |
pat 2 |
| b19 |
2.517 |
1.607 |
0.820 |
0.365 |
0.006 |
0.110 |
16.92Å |
pat 3 |
| b20 |
2.446 |
1.578 |
0.802 |
0.334 |
0.005 |
0.103 |
17.53Å |
NEW BEST |
| b21 |
2.482 |
1.589 |
0.797 |
0.362 |
0.006 |
0.104 |
17.54Å |
pat 1 |
| b22 |
2.488 |
1.607 |
0.801 |
0.347 |
0.005 |
0.107 |
17.42Å |
pat 2 |
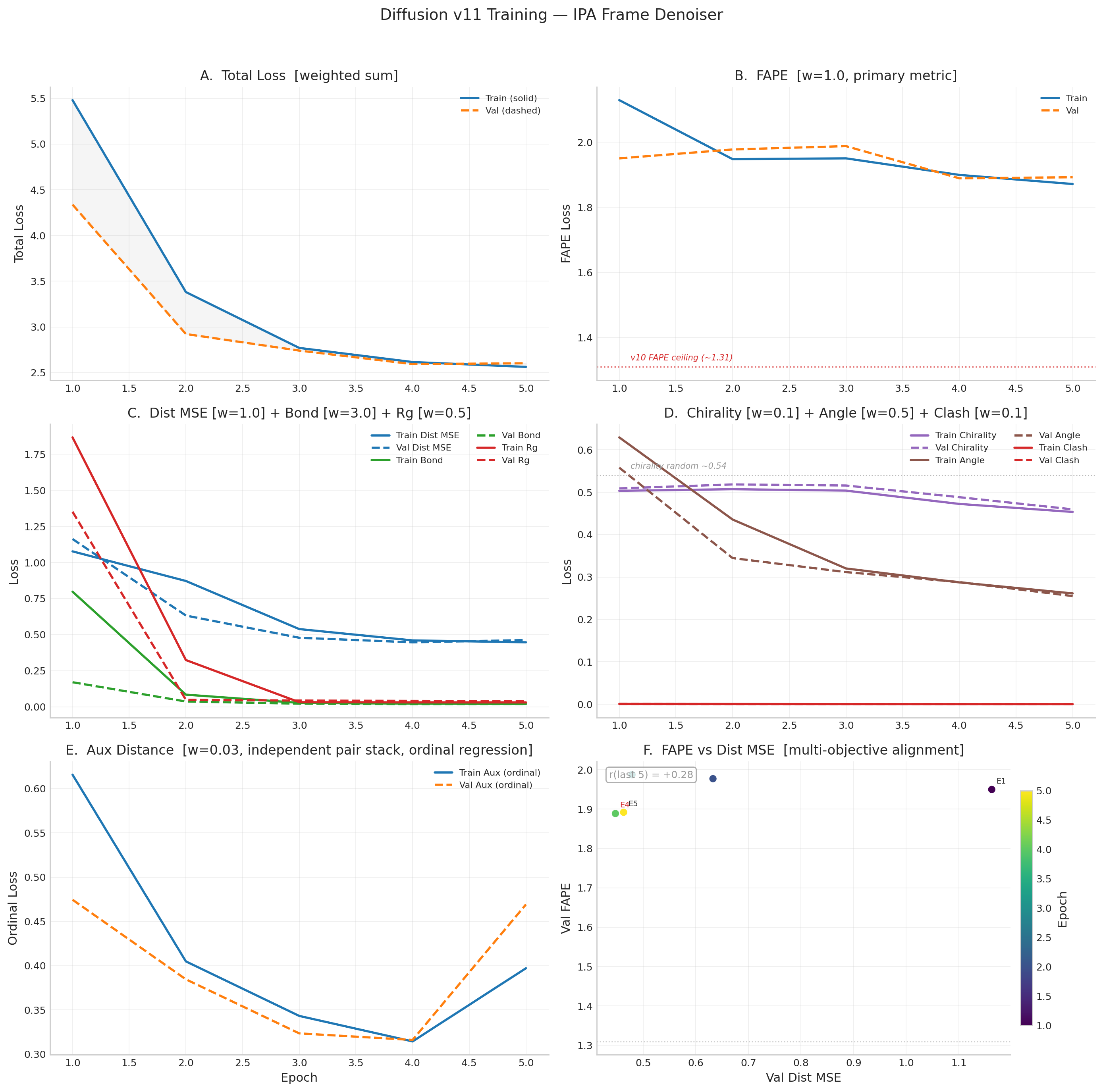
v11b Historical Results (14 epochs, gradient competition collapse at E8 — superseded by v12)
v11b validated the IPA + frame rotation loss design. Best epoch E8: FAPE 1.655, frame_rot 0.830, TM 0.093.
After E8, gradient competition between dist_mse and frame_rot through shared 128-dim single representation
caused collapse — TM dropped to 0.061, frame_rot reverted to 0.926. Stopped at E14. v12 addresses this
with 4.1x capacity (31.1M params, d_ipa_hidden=512, 2-layer FrameUpdate MLP).
v11b Training Table (E1–E14)
| Epoch | Val Total | Val FAPE | Frame Rot | Val Dist | Val Bond | DDIM TM | Status |
| 1 | 3.057 | 1.934 | 1.067 | 0.409 | 0.015 | 0.094 | BEST |
| 5 | 2.736 | 1.756 | 0.888 | 0.384 | 0.010 | 0.091 | BEST |
| 8 | 2.580 | 1.655 | 0.830 | 0.351 | 0.009 | 0.093 | BEST (peak) |
| 11 | 2.797 | 1.813 | 0.880 | 0.374 | 0.008 | 0.079 | pat 3 |
| 14 | 2.973 | 1.898 | 0.926 | 0.423 | 0.011 | 0.061 | pat 6 (stopped) |
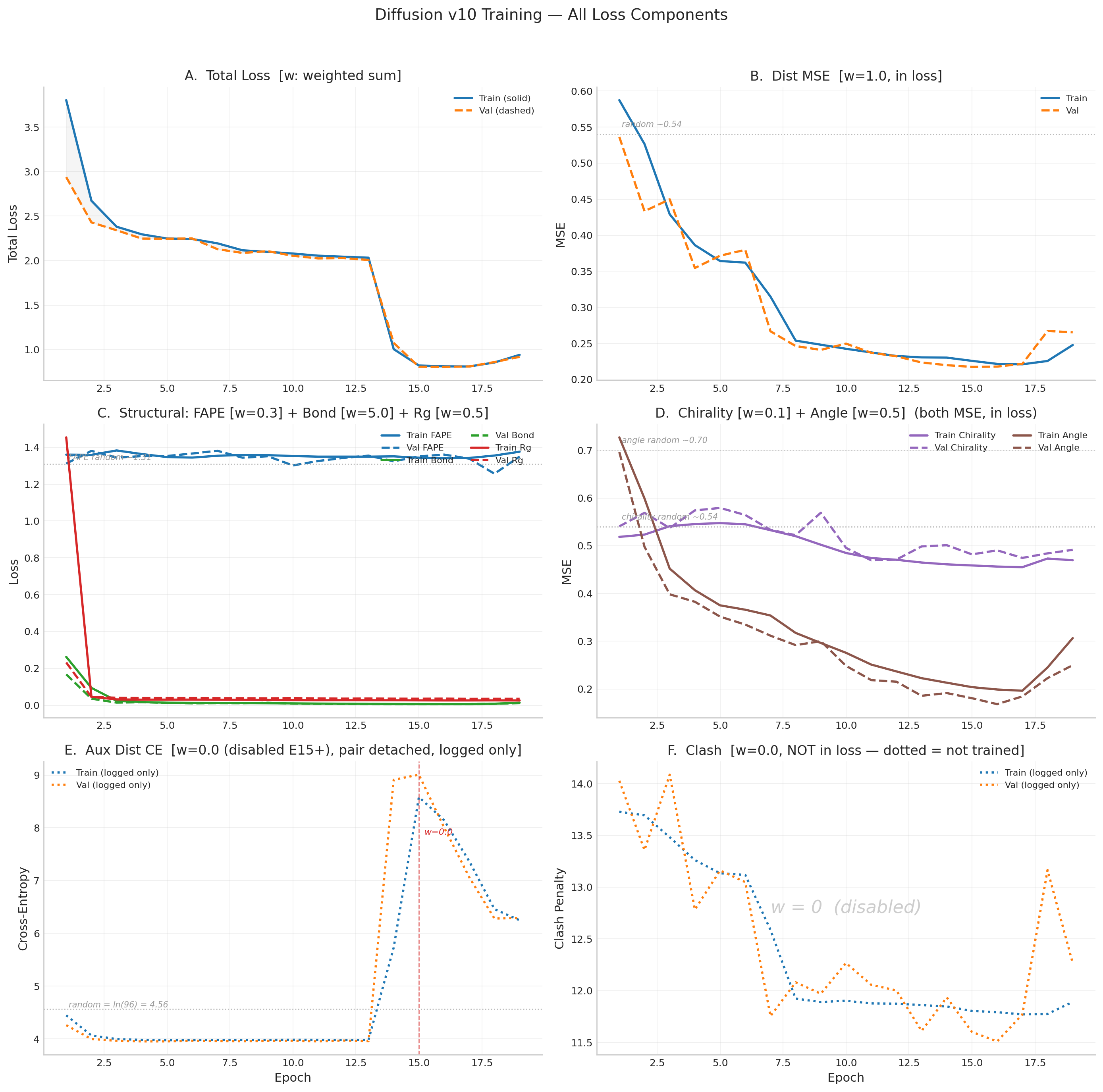
v11 Historical Results (15 epochs, no frame rotation supervision — superseded)
v11 used the same IPA architecture but had a critical bug: x0_pred = t_vec discarded
learned rotation matrices R. FAPE rebuilt frames from coordinates via Gram-Schmidt, so rotation
quaternions received no direct gradient. FAPE peaked at 1.818 (E10) then degraded to 2.052 (E15).
DDIM TM-score collapsed from 0.095 to 0.046. Stopped after E15 for architectural fix.
v11 Loss Curves
v11 Training Table (E1–E15)
| Epoch | Val Total | Val FAPE | Val Dist | Val Bond | DDIM TM | Status |
| 1 | 4.338 | 1.950 | 1.163 | 0.171 | 0.099 | BEST |
| 3 | 2.741 | 1.988 | 0.479 | 0.022 | 0.128 | BEST |
| 6 | 2.558 | 1.865 | 0.448 | 0.019 | 0.095 | BEST |
| 10 | 2.431 | 1.818 | 0.382 | 0.015 | 0.087 | BEST (peak) |
| 13 | 2.539 | 1.919 | 0.380 | 0.016 | 0.053 | pat 3 |
| 15 | 2.726 | 2.052 | 0.411 | 0.019 | 0.046 | pat 5 (stopped) |
Loss Reference & Targets
| Metric |
w |
Type |
v11b E1 |
v11b E5 |
v12 E1 |
v12 E10 |
Target / Interpretation |
| FAPE |
1.0 |
L1 |
1.934 |
1.756 |
2.074 |
1.651 |
Primary metric. Uses learned R_pred frames. v10 ceiling=1.31, untrained >2.0. <1.0 = correct folds. |
| Frame Rot |
0.5 |
1−cosθ |
1.067 |
0.888 |
1.199 |
0.833 |
Angular error of learned R vs Gram-Schmidt truth. 1.0 = ~90° (random), 0 = perfect. Target: <0.5 by E15. |
| Dist MSE |
1.0 |
MSE |
0.409 |
0.384 |
0.857 |
0.352 |
Pairwise Cα distance error. <0.1 = sub-Å accuracy. Plateauing ~0.35–0.40. |
| Bond |
3.0* |
MSE |
0.015 |
0.010 |
0.135 |
0.008 |
Cα–Cα bond error. *annealed 1→3. Solved. |
| Rg |
0.5 |
MSE |
0.038 |
0.038 |
1.805 |
0.038 |
Radius of gyration error. Converged by E3. |
| Chirality |
0.1 |
MSE |
0.428 |
0.369 |
— |
— |
Backbone handedness. Random ~0.54. Improving. |
| Angle |
0.5 |
MSE |
0.202 |
0.154 |
— |
— |
Cα bond angles. Random ~0.70. Well below random. |
| Clash |
0.1 |
penalty |
0.0002 |
0.0001 |
— |
— |
Steric clashes (3.8Å threshold). Essentially zero. |
| Aux Ordinal |
0.03 |
ordinal |
0.322 |
0.316 |
— |
— |
Independent pair stack + ordinal regression. Stable. |
| TM-score |
— |
DDIM |
0.094 |
0.091 |
0.101 |
0.102 |
50-step DDIM. Target: >0.15 by E10, >0.30 by E20. >0.17 = recognizable folds. E2 peak=0.132. |
| RMSD |
— |
DDIM |
17.22Å |
17.62Å |
15.28Å |
17.09Å |
<10Å = partial fold. <5Å = high quality. E2 best=13.73Å. DDIM flat ~17Å since E4. |
v12b Training — b20 NEW BEST, consolidating at b21-b22
- b20 remains best: val_total 2.446, val FAPE 1.578, frame_rot 0.802. b21-b22 show mild patience (pat 2/15) with val_total 2.48-2.49, close to best. Train loss still declining (2.52→2.52), no signs of overfitting.
- Pattern: consolidate then breakthrough. b14→b16 gave 3 consecutive bests, b17-b19 consolidated (pat 3), b20 broke through. Now b21-b22 consolidating again (pat 2). Expecting another push soon. LR at 1.49e-5 (cosine decay), plenty of schedule remaining.
- v12 E11-E13 regression diagnosed: Gradient starvation via global
clip_grad_norm_(1.0). Denoiser (28.5M params) consumed 99%+ of gradient budget. pair_stack_norm collapsed mid-E12: 0.125→0.007→0.000.
- Gradient cosine diagnostic confirmed: No gradient competition. FAPE & frame_rot are aligned (+0.57 cosine). dist_mse is near-orthogonal to both (+0.14). Losses do not oppose each other — the regression was purely gradient starvation, not conflicting objectives.
- v12b fixes applied (rolling back to E10 EMA weights):
- Per-module gradient clipping: denoiser max_norm=1.0, pair_stack=0.5, aux_pair=0.5
- Per-module LR groups: pair_stack gets 3x base LR (6e-5) to compensate gradient attenuation
- Pair stack tripwire: if grad norm < 0.01 for 50 consecutive steps, inject gradient noise (scale=0.01)
- Hard halt: if grad norm < 0.001 for 500 total steps, stop training
- Atomic checkpoint saves (write to tmp file, then rename) to survive SLURM preemption
- EMA step counter preserved across checkpoint resumes
- NaN loss batches skipped entirely instead of poisoning gradient accumulator
- Architecture bugs fixed by code review:
- Frame masking: was zeroing rotation matrices (degenerate non-SO(3)), now uses identity fallback
slerp_to_identity: added large-angle guard for θ near π (sin→0 blow-up)- Gradient checkpointing: IPA layers use
use_reentrant=True (required for AMP; False causes metadata mismatch)
DiffusionSchedule tensors registered as buffers (no more manual .to(device))
Gradient cosine similarity diagnostic (E10 vs E12 vs E13)
Per-loss gradient directions on shared parameters (denoiser + pair_stack). Negative cosine = direct competition. Positive = aligned.
| Loss Pair |
E10 (best) |
E12 |
E13 |
Verdict |
| FAPE vs frame_rot |
+0.59 |
+0.57 |
+0.50 |
Aligned |
| dist_mse vs FAPE |
+0.16 |
+0.14 |
+0.41 |
Near-orthogonal |
| dist_mse vs frame_rot |
+0.24 |
+0.00 |
-0.02 |
Near-orthogonal |
| FAPE vs bond_geom |
-0.04 |
-0.23 |
+0.02 |
Near-orthogonal |
Key finding: No gradient competition between any loss pair. FAPE and frame_rot are strongly aligned (+0.5 to +0.6), confirming they learn consistent frame representations. The regression was caused entirely by gradient starvation of the pair_stack module — not conflicting loss objectives.
v11 post-mortem (different failure mode)
| Hypothesis |
Result |
| Frame confidence starving updates |
NO — 57.7% conf > 0.5 |
| Gradients dead at frame_update |
NO — highest grad norm (4.017) |
| FAPE gradient reaches frame_update |
YES — 13.03 (largest) |
| Learned R used in output |
NO — R discarded |
Root cause (v11): x0_pred = t_vec discarded learned R. v11b fix: frame_rotation_loss + fape_loss_with_frames using learned R.
Last updated: 2026-03-10 21:07 UTC
v10 Historical Results (21 epochs, EGNN — superseded)
v10 used an 8-layer EGNN denoiser (14.6M params). After 21 epochs: dist_mse 60% below random (0.218),
bond essentially solved (0.006), but FAPE stuck at random (~1.31) and TM-score peaked at 0.131.
Two aux_dist_ce divergence incidents required intervention (pair detach at E5, full disable at E15).
Cosine LR restarts caused pathological degradation at the minimum.
Best val structural = 0.805 (E16). DDIM best: TM=0.131, RMSD=14.53Å.
Diffusion v11 — IPA-Based Frame Denoising (Current)
Why the Pivot from v10
v10 used an 8-layer EGNN denoiser that updates Cα coordinates through distance-weighted
pairwise messages. After 21 epochs on CATH, it achieved dist_mse 60% below random
(learning “proteins are compact blobs of the right size”) but FAPE remained at
its random baseline (~1.31) and TM-score peaked at 0.131 (random ~0.10). The model could not
learn correct protein topology.
Root cause: EGNN has no concept of local reference frames. It passes messages
based on pairwise distances and updates coordinates through distance-weighted vectors. FAPE
measures frame-aligned point error — whether predicted local coordinate frames match ground
truth frames — which EGNN has no inductive bias to optimize. Increasing w_fape on the old
architecture means fighting the inductive bias.
v11 Overview
v11 replaces the EGNN denoiser with an Invariant Point Attention (IPA) structure
module that explicitly maintains and refines per-residue rigid-body frames (rotation ∈ SO(3)
+ translation ∈ ℝ³). The architecture class is the same used in AlphaFold2’s
structure module and RFDiffusion. Everything upstream (frozen ContactClassifier encoder, pair stack,
Rg predictor) is carried forward from v10 — these components work.
8.4M total parameters (7.2M trainable, 1.2M frozen encoder). Training on CATH (18,024 train /
608 val, max 125 residues) on a single A40 GPU.
IPA Denoiser (6 layers)
Each IPA block performs three operations:
- Invariant Point Attention — standard multi-head attention on the single
representation, augmented with (a) pair bias from the contact-conditioned pair stack and
(b) 3D point attention: each head generates query/key/value points in ℝ³ that are
transformed into each residue’s local frame. Attention weights depend on geometric
distances between learned points — invariant to global rotation/translation.
- Transition MLP — 2-layer feedforward on the single representation.
- Frame update — predicts a small quaternion + translation update per residue,
composed in the local frame (right-multiplication for SE(3) equivariance). Initialized near-zero
so frames are approximately preserved in early training.
The pair representation is static through the IPA stack (not updated). If FAPE stalls
after E15, adding outer-product pair updates is the first planned intervention.
SNR-Gated Frame Initialization
Per-residue rigid frames are built from noised Cα coordinates via Gram-Schmidt
orthogonalization on consecutive backbone triplets. At high noise (σt where
SNR < 1.0, roughly t > 700), the noised coordinates are near-isotropic and Gram-Schmidt
is numerically unstable. The frame confidence mechanism smoothly blends toward identity frames:
$$\text{conf}(t) = \text{clamp}\!\left(\frac{\text{SNR}(t) - 0.2}{1.0 - 0.2},\, 0,\, 1\right), \qquad
R_{\text{init}} = \text{slerp}(I,\, R_{\text{GS}},\, \text{conf})$$
The first IPA layer’s frame update is scaled by this confidence, so unreliable initial
frames at high noise are attenuated rather than propagated.
Frame-Aware Self-Conditioning
50% of training steps: run a no-grad forward pass to get x0prev, build clean
frames from it (treated as t=0), and use those as the initial frames for the second pass. At high
noise where xt frames are identity, self-conditioning provides the model’s best
guess at clean local geometry — the IPA layers refine good frames instead of building them
from scratch. This is qualitatively more powerful than v10’s coordinate-only self-conditioning.
Fixed-Scale Coordinates
v11 replaces per-protein Rg normalization with division by a fixed constant (10Å).
Why: Rg normalization caused the noise schedule to be protein-size-dependent.
A protein with Rg=5Å had coordinate values ~1.0 after normalization while Rg=25Å gave
~0.2–0.5, meaning the same noise level destroyed more signal for larger proteins. This
silently caps TM-scores and looks like a “plateau” rather than a systematic bias.
All successful protein diffusion models (FrameDiff, RFDiffusion, Genie) use fixed-scale coordinates.
Independent Auxiliary Distance Head
v10’s DistanceHead read from the main pair stack through detach(), causing
feature drift divergence (aux_dist_ce: 3.95→38.8 over 3 epochs). v11 uses a completely
independent lightweight pair stack (64-dim, 2 triangle attention layers, ~500K params) reading from
the frozen encoder outputs. Ordinal regression with 32 bins replaces 96-bin
cross-entropy — adjacent-bin errors are penalized proportionally to distance, not as hard
misclassifications.
v11 Loss Weights
| Loss |
v10 |
v11 |
Change rationale |
| FAPE | 0.3 | 1.0 | IPA can actually optimize frame consistency |
| Bond | 5.0 | 3.0 | Gentler anneal (1→3 over E1-E10), avoids tug-of-war |
| Clash | 0.0 | 0.1 | Re-enabled: fixed-scale coords make 3.8Å threshold meaningful |
| Aux dist | 0.0 | 0.03 | Conservative start for new independent pair stack |
| Dist MSE | 1.0 | 1.0 | — |
| Angle | 0.5 | 0.5 | — |
| Chirality | 0.1 | 0.1 | — |
| Rg | 0.5 | 0.5 | — |
v11 Training Protocol
LR: 5e-5 (halved from v10). Cosine decay to 1e-6 over 55 epochs with 3-epoch warmup, no
restarts (v10 restarts caused pathological degradation at the LR minimum). 1000 diffusion
timesteps (was 200). DDIM-50 evaluation every epoch. DDPM training with cosine noise schedule.
Phase 1 (E1-E10): FAPE must be ≥10% below E1 baseline by E5, ≥20% by E10.
Phase 2 (E10-E40): Target TM-score > 0.3.
Phase 3 (E40-E60): LR decay for final refinement and EMA stabilization.
Diffusion v10 — Architecture & Loss Function (Previous)
Overview
We train a denoising diffusion model for protein backbone (Cα) structure generation,
conditioned on inter-residue contact maps predicted by a frozen ContactClassifier encoder.
The model operates in Rg-normalized coordinate space: all coordinates are divided by
the radius of gyration so the diffusion process is scale-invariant. The denoiser is an
8-layer SE(3)-equivariant graph neural network (EGNN) with 14.6M parameters (13.4M trainable,
1.2M frozen encoder). Training uses the CATH dataset (18,024 train / 608 val proteins,
max 125 residues).
Total Loss
The total loss is a weighted combination of eight components. At epoch \(e\):
$$\mathcal{L}_{\text{total}} = w_{\text{dist}} \cdot \mathcal{L}_{\text{dist}} + \beta(e) \cdot \mathcal{L}_{\text{bond}} + w_{\text{aux}} \cdot \mathcal{L}_{\text{aux}} + w_{\chi} \cdot \mathcal{L}_{\chi} + w_{\theta} \cdot \mathcal{L}_{\theta} + w_{\text{fape}} \cdot \mathcal{L}_{\text{fape}} + w_{\text{rg}} \cdot \mathcal{L}_{\text{rg}}$$
Clash loss and auxiliary distance CE are logged but excluded (\(w_{\text{clash}} = 0\), \(w_{\text{aux}} = 0\)). See incident reports for rationale.
1. Distance MSE \(w_{\text{dist}} = 1.0\)
Mean squared error on all pairwise Cα distances in Rg-normalized space:
$$\mathcal{L}_{\text{dist}} = \frac{1}{|\mathcal{M}|} \sum_{(i,j) \in \mathcal{M}} \left( \| \hat{x}_i^{(0)} - \hat{x}_j^{(0)} \| - \| x_i^{(0)} - x_j^{(0)} \| \right)^2$$
where \(\hat{x}^{(0)}\) is the predicted clean structure, \(x^{(0)}\) is the ground truth, both in
Rg-normalized coordinates, and \(\mathcal{M}\) is the set of valid residue pairs. Clamped to max 10.0.
Random baseline: ~0.54 (MSE of Gaussian noise pairwise distances vs true).
2. Bond Geometry \(\beta(e) = \min(5.0,\; 1.0 + 4.0 \cdot \min(e/15, 1))\)
MSE on consecutive Cα–Cα distances against the ideal 3.8Å bond length (in Rg-normalized space):
$$\mathcal{L}_{\text{bond}} = \frac{1}{L-1} \sum_{i=1}^{L-1} \left( \| \hat{x}_i^{(0)} - \hat{x}_{i+1}^{(0)} \| - \frac{3.8}{R_g} \right)^2$$
The weight is annealed from 1.0 to 5.0 over the first 15 epochs. Starting low prevents bond geometry
from dominating early training when the model hasn't learned global structure. As training matures, the
increasing weight enforces physically valid backbone geometry. Clamped to max 10.0.
Random baseline: ~0.17. Below 0.02 indicates bonds are within 0.1Å of the ideal 3.8Å spacing.
3. FAPE (Frame-Aligned Point Error) \(w_{\text{fape}} = 0.3\)
Measures local structural consistency by constructing rigid frames from consecutive Cα triplets
and computing the error in each frame's local coordinate system:
$$\mathcal{L}_{\text{fape}} = \frac{1}{N_f \cdot L} \sum_{f=1}^{N_f} \sum_{j=1}^{L} \min\!\Big( \| R_f^{\top}(\hat{x}_j - o_f) - R_f^{*\top}(x_j - o_f^*) \|,\; d_{\text{clamp}} \Big)$$
Frames are built from every other triplet of Cα atoms: the x-axis along \(c_2 - c_0\), z-axis from the
cross product, y-axis completing the right-handed system. Unlike AlphaFold's random 14-residue sampling,
v10 uses all residues as targets for stable gradients. Clamped at \(d_{\text{clamp}} = 10.0\).
Random baseline: ~1.31. Drops below 1.0 when the model learns correct fold topology.
This is the hardest loss to reduce because it requires global structural correctness, not just local geometry.
4. Radius of Gyration \(w_{\text{rg}} = 0.5\)
MSE on log-transformed radius of gyration predictions for scale invariance:
$$\mathcal{L}_{\text{rg}} = \left( \log \hat{R}_g - \log R_g \right)^2$$
Since the denoiser works in Rg-normalized space, a separate MLP (\(\texttt{RgPredictor}\))
predicts the absolute radius of gyration from sequence embeddings. This allows
recovering real-space coordinates at inference: \(x_{\text{real}} = \hat{R}_g \cdot \hat{x}_{\text{norm}}\).
Random baseline: ~0.23. Converges below 0.05 by E2.
5. Chirality \(w_{\chi} = 0.1\)
MSE on normalized signed volumes (scalar triple products) of consecutive Cα quartets,
ensuring correct backbone handedness:
$$\mathcal{L}_{\chi} = \frac{1}{L-3} \sum_{i=1}^{L-3} \left( \frac{\mathbf{v}_1 \cdot (\mathbf{v}_2 \times \mathbf{v}_3)}{\|\mathbf{v}_1\| \|\mathbf{v}_2\| \|\mathbf{v}_3\|} \bigg|_{\hat{x}} - \frac{\mathbf{v}_1 \cdot (\mathbf{v}_2 \times \mathbf{v}_3)}{\|\mathbf{v}_1\| \|\mathbf{v}_2\| \|\mathbf{v}_3\|} \bigg|_{x} \right)^2$$
where \(\mathbf{v}_1 = x_{i+1} - x_i\), \(\mathbf{v}_2 = x_{i+2} - x_{i+1}\), \(\mathbf{v}_3 = x_{i+3} - x_{i+2}\).
The normalization maps volumes to \([-1, 1]\), making the loss scale-invariant.
Natural proteins are L-amino acids with consistent chirality; this loss prevents mirror-image structures.
Random baseline: ~0.54 (expected MSE for random values in [-1, 1]).
6. Bond Angle \(w_{\theta} = 0.5\)
MSE on cosines of Cα–Cα–Cα bond angles for consecutive triplets:
$$\mathcal{L}_{\theta} = \frac{1}{L-2} \sum_{i=1}^{L-2} \left( \cos\hat{\theta}_i - \cos\theta_i \right)^2, \quad \cos\theta_i = \frac{(\hat{x}_{i+1} - \hat{x}_i) \cdot (\hat{x}_{i+2} - \hat{x}_{i+1})}{\| \hat{x}_{i+1} - \hat{x}_i \| \| \hat{x}_{i+2} - \hat{x}_{i+1} \|}$$
The ideal Cα–Cα–Cα angle in proteins is ~120° (\(\cos\theta \approx -0.5\)).
Working in cosine space avoids discontinuities at 0°/360°.
Random baseline: ~0.70. Below 0.1 indicates correct backbone angles.
7. Auxiliary Distance Cross-Entropy \(w_{\text{aux}} = 0.0\) (disabled)
Cross-entropy loss on binned pairwise distances, predicted by a DistanceHead MLP from the
detached pair representation:
$$\mathcal{L}_{\text{aux}} = -\frac{1}{|\mathcal{M}'|} \sum_{(i,j) \in \mathcal{M}'} \log p_{ij}\big[\text{bin}(d_{ij})\big], \quad \text{bin}(d) = \left\lfloor \frac{d - d_{\min}}{\Delta} \right\rfloor, \quad \Delta = \frac{d_{\max} - d_{\min}}{N_{\text{bins}}}$$
Parameters: \(N_{\text{bins}} = 96\), \(d_{\min} = 2\)Å, \(d_{\max} = 40\)Å, \(\Delta = 0.396\)Å/bin.
Distances are computed in real space (\(d_{ij} = R_g \cdot \|x_i - x_j\|\)) then binned.
Only pairs within \(d_{\max}\) are included.
The pair representation is detached before entering the DistanceHead, meaning aux gradients
train only the MLP head, not the pair stack. This is critical — an earlier experiment without detach
caused exponential divergence (see incident report in Training tab).
Random baseline: \(\ln(96) = 4.56\). A uniform predictor assigns \(1/96\) to each bin,
giving \(-\ln(1/96) = \ln(96)\). Weight history: 0.3 (E1–E13) → 0.03 (E14) → 0.0 (E15+, disabled).
Disabled because the pair representation evolves via structural losses while the DistanceHead observes it
through a stop-gradient wall, causing irrecoverable feature drift and exponential CE divergence.
Still computed and logged as a diagnostic.
8. Clash Loss \(w_{\text{clash}} = 0.0\) (disabled)
Contact-guided steric clash penalty on non-bonded atoms closer than 3.0Å:
$$\mathcal{L}_{\text{clash}} = \frac{1}{|\mathcal{N}|} \sum_{(i,j) \in \mathcal{N}} \left[ (1 + 2 \cdot c_{ij}) \cdot \text{ReLU}(3.0 - d_{ij}) \right]^2$$
where \(\mathcal{N}\) is the set of non-bonded pairs (\(|i - j| > 1\)), \(c_{ij}\) is the predicted
contact probability (detached), and \(d_{ij}\) is the Rg-normalized distance.
Why disabled: The 3.0Å threshold is applied in Rg-normalized coordinates, but for a
typical protein with \(R_g \approx 10\)Å, this maps to \(3.0 \times 10 = 30\)Å in real space —
penalizing nearly all non-bonded pairs. This produced noisy gradients that dominated ~47% of the total
loss in v9, drowning the structural learning signal. Removing clash led to immediate training stability
and the structural losses handle steric quality implicitly (clash decreases organically as the model
generates better structures).
Training Configuration
| Optimizer | AdamW (\(\beta_1=0.9, \beta_2=0.999\)) |
| Peak learning rate | \(10^{-4}\) |
| LR schedule | 5-epoch linear warmup (\(0.01 \times \text{lr} \to \text{lr}\)), then CosineAnnealingWarmRestarts (\(T_0 = 15\) epochs, \(\eta_{\min} = 10^{-5}\)) |
| Batch size | 8 (grad accumulation = 2, effective = 16) |
| Mixed precision | AMP with GradScaler, \(\texttt{dx\_clamp} = 0.5\) |
| EMA | Decay = 0.999, used for DDIM evaluation |
| Self-conditioning | 50% probability during training |
| Early stopping | Patience = 15 epochs on val structural loss (excludes aux_dist_ce) |
| DDIM evaluation | 50 steps, every 3 epochs, using EMA weights |
| Hardware | Single NVIDIA A40 (48 GB), ~15 min/epoch |
| Dataset | CATH 4.3: 18,024 train / 608 val, max 125 residues |
DDIM Evaluation Metrics
Every 3 epochs, we generate structures via 50-step DDIM sampling using EMA weights and evaluate against
ground truth:
- TM-score (Template Modeling): Global fold similarity, range [0, 1]. Scores > 0.17 indicate
the same fold; > 0.5 is the same topology. Random structures score ~0.10.
- RMSD (Root Mean Square Deviation): Average atomic displacement after optimal superposition.
Random placement gives ~15–16Å. Below 5Å is high quality.
- GDT (Global Distance Test): Fraction of residues within 1–8Å of the true position.
Random gives ~3–4%.